Typing Sanskrit can be challenging if you don’t have access to your special keyboard, or don’t have your favorite input tools installed on the computer you are working on.
So I set out to write a Google Docs add-on that could make it easy to do so. A Google Docs add-on could be an ideal option for typing Sanskrit, even while using guest computers.
Sanskrit has more sounds than English and other languages written in Roman scripts. Devanagari is the commonly used script used for writing Sanskrit and uses non-ASCII characters. Some systems such as IAST and ITRANS use additional symbols to represent Sanskrit sounds in Roman-like scripts. For example, ā in IAST is used to represent the long “a” syllable.
I found this library called Sanscript.js which could convert between these different writing schemes. However it doesn’t address the problem of being able to use a standard keyboard with ASCII characters. IAST includes additional characters such as ū, ṣ, ñ, ṅ, ṃ, etc. It is more readable than other competing schemes such as ITRANS. ITRANS includes a mix of upper-case and lower-case letters, in the middle of a word, to represent additional sounds. This has adverse affect on the aesthetics of the script. I find it harder to read as well. IAST has also been used in academia and Sanskrit books written in the West since a long time. As a result many users of Sanskrit language are familiar with this system. It was later standardized as ISO 15919 with small changes.
I added a new “IAST Simplified” scheme to the list supported by Sanscript.js. This is inspired from my favorite Sanskrit writing software called Sanskrit Writer. This scheme uses standard ASCII characters and closely resembles the IAST scheme. For example, it uses –a for ā, ~n for ñ, and h. for ḥ, and so on. The following table explains this scheme in detail:
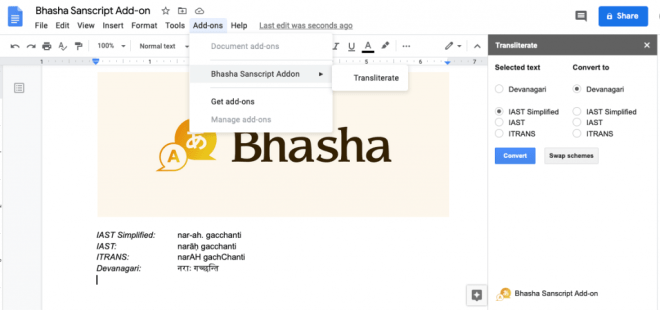
After these additions to Sanscript.js, I was able to write a quick Google Docs plugin that could convert between different schemes with a click of a button. You can try this plugin out by visiting this link, or searching for “Bhasha” in Google Docs under addons.
I was hoping to have a WYSIWYG design where transliteration could happen as you typed. This required intercepting each edit, and was not possible using the Google Docs API. As a second option, I wrote a plugin for QuillJS, a cool open-source rich-text editor. You can try out this add-on here: https://trivedigaurav.com/exp/bhasha/.

This may not be mobile friendly. I didn’t spend time test it on the phone since you can easily switch to a Sanskrit keyboard on a phone anyway.
Source code for the QuillJS plugin is on Github here. Please feel free to hack on it!
Edit 1: Bhasha is now available as a Google Docs add-on.
Edit 2: Added Vedic accents!
IAST: a॒gnimī॑ḻe pu॒rohi॑taṃ ya॑jñasya॑ de॒vamṛ॒tvija॑m
Devanagari: अ॒ग्निमी॑ळे पु॒रोहि॑तं य॑ज्ञस्य॑ दे॒वमृ॒त्विज॑म्
IAST Simplified:a\_gnim-i\!~le pu\_rohi\!tam. ya\!j~nasya\! de\_vamr.\_tvija\!m
Edit 3: Added Double Tone Svarita!
Devanagari: स्थि॒रैरङ्गै᳚स्तुष्टुवाग्ँस॑स्त॒नूभिः॑
IAST Simplified: sthi\_raira.ngai\=stus.t.uv-ag~csa\!sta\_n-ubhih.\!

